Part 1: A deep dive into multi-database architecture, AI-powered entity extraction, and intelligent document processing
Introduction
Traditional RAG systems rely solely on vector embeddings for semantic search, but this approach has fundamental limitations. As I discussed in my earlier post “Beyond Vector Search: How GraphRAG Enables Smarter AI Responses”, vector-based systems struggle with entity-focused queries like “What did Microsoft say about AI safety?” or “Show me all papers by authors who work on neural architecture search” because they lack understanding of relationships between concepts, organizations, and people.
GraphRAG addresses these limitations by combining knowledge graphs with vector embeddings and thus, creating a document intelligence system that understands both semantic similarity and structural relationships.
This is Part 1 of a multi-part blog that will document development of a GraphRAG document repository server built to support AI agents or a Chat application that require retrieval of additional information from repository corpus to perform an actions or generate response. Users can upload a PDF file or submit a web page by URL to create a document that is then undergoes a series of automated steps in the ingestion pipeline – token-based chunking, named entities and entity relationships extraction, embeddings generation. Resulting objects are store in a multi-database architecture that provides both semantic and graph-based search capabilities.
The Part 1 focus is on architectural decisions made and approaches taken to develop document ingestion pipeline and AI-powered named entities and relationships extraction that provide the foundation layer of the GraphRAG system back-end.
System Architecture Overview
The most critical architectural decision was using three specialized databases instead of forcing everything into a single system. Each database is optimized for a specific type of data and query pattern.
High-Level Architecture

Multi-Database Strategy
Neo4j (Knowledge Graph)
- Purpose: Store documents, text chunks, entities, and authors as graph nodes
- Strengths: Relationship traversal, pattern matching, entity connections
- Query Types: “Find documents by authors who also wrote about X”, “Show entity relationships”
- Why: Graph databases excel at relationship-heavy queries that would require complex JOINs in SQL
ChromaDB (Vector Database)
- Purpose: Store high-dimensional embeddings (1024 dimensions) for semantic search
- Strengths: Fast approximate nearest neighbor search, semantic similarity
- Query Types: “Find conceptually similar content”, semantic search
- Why: Specialized vector databases provide orders of magnitude better performance than storing vectors in general-purpose databases
MinIO (Object Storage)
- Purpose: Store original files, extracted metadata, processing logs
- Strengths: Scalable file storage, S3-compatible API, presigned URLs
- Query Types: Direct file access, metadata retrieval
- Why: Object storage is purpose-built for files, with features like versioning and presigned URLs
Why Not a Single Database?
PostgreSQL with pgvector: Good for smaller projects but graph traversal becomes complex and slow at scale. SQL JOINs can’t match Neo4j’s native graph traversal performance.
Neo4j alone: Can store vectors as node properties, but vector similarity search is significantly slower than specialized vector databases like ChromaDB or Pinecone.
Vector database alone: Excellent for semantic search but can’t efficiently answer relationship queries like “documents by authors who cite this work” or build entity networks.
The three-database approach provides the best tool for each job. While it adds operational complexity, the performance gains and query flexibility justify the investment.
Design Philosophy
Separation of Concerns: Each service layer has a single, well-defined responsibility. Processing services orchestrate workflows, extraction services handle content, AI services integrate external APIs, and graph services manage database operations.
Service-Oriented Architecture: Services communicate through well-defined interfaces, making it easy to swap implementations (e.g., replace Claude with another LLM, swap ChromaDB for Pinecone).
Async Operations: Document processing involves I/O-heavy operations (API calls, database writes). Asynchronous processing enables concurrent operations and better resource utilization.
Dependency Injection: Database clients and services are injected as dependencies, facilitating testing and enabling different configurations for development vs. production.
Current Technology Stack
REST API Framework – FastAPI is a modern, fast (high-performance), web framework for building APIs with Python based on standard Python type hints.
- Modern async web framework with automatic API documentation
- Native support for async / await patterns
- Type safety with Pydantic models (Pydantic is a data validation library for Python)
Graph Database – Neo4j Community Edition a high-performance graph database
- Industry-leading graph database with Cypher query language
- ACID transactions for data consistency
- Flexible schema evolution
Vector Database – Chroma, an open-source vector database with vector, full-text, regex, and metadata search support
- Lightweight wrappers around popular embedding providers like OpenAPI or Cohere
Object storageas a shared layer forquery nodeswhich resolve user queriesCompactor nodes, which asynchronously build indexes and persist them to object storage- Good performance for development and medium-scale production
Object Storage – MinIO, a high-performance S3-compatible object store
- MinIO AIStor contains every component required to run large scale data infrastructure
- Self-hosted for development
- Allows easy migration to cloud S3
AI Services:
- Anthropic Claude 3.5 Haiku: Entity extraction with structured outputs (~100ms per chunk)
- VoyageAI voyage-3: High-quality 1024-dimensional embeddings
- VoyageAI rerank-2: Result re-ranking
Processing Libraries:
- PyMuPDF: Fast PDF text extraction
- BeautifulSoup4: HTML parsing and web scraping
- tiktoken: Token-aware text chunking
Document Processing Pipeline
At present, you can add a new document to the repository by uploading a PDF file to MinIO object storage or by submitting a web page URL. The document processing pipeline transforms content of raw files and web pages into a rich knowledge graph with semantic embeddings. The pipeline consists of seven stages:
- File upload / Web page content scrapping
- Plain text extraction from PDF file or page content
- Token-aware text chunking (max chunk size is 1000 tokens with 15% overlap)
- Named entity extraction from documents (Author, Person, Organization, Location, Technology, Concept, Product and Event)
- Knowledge Graph update (add new Document node and its Chunk nodes; add Author nodes and Entity nodes of Person; finally, add edges to define relevant relationships between new and existing nodes).
- Generate embedding vectors for plain text stored for each Chunk node and the store embeddings in vector DB (Chroma)
Stage 1: Add New Document to the Repository
Currently, GraphRAG document repository supports two options for adding new documents:
Upload a PDF file (multipart file upload):
- Calculate MD5 hash for de-duplication and check if document already exists
- Store new binary file in MinIO object storage
- Generate unique document ID
Submit a web page URL:
- Validate URL format and fetch content with appropriate headers
- Archive HTML content and store it in MinIO object storage
- Generate unique document ID
Key Considerations for adopting this approach:
- De-duplication: Hash-based detection prevents processing the same document twice
- File Validation: Size limits (e.g., 50MB) and format checks prevent malicious content
- Unique IDs: UUIDs ensure globally unique document identifiers
Stage 2: Plain Text Extraction from Documents
Next step after new document is added to repository, is the plain text extraction from it to allow named entity extraction (NPE) from it and chunking for generating embeddings.
Text extraction from a PDF file (PyMuPDF – a Python library for working with PDF files):
- Extract text from all pages preserving document structure where possible
- Extract PDF metadata (such as author, title, creation date, page count)
- Handle various PDF versions and encodings
- Error handling for corrupted or password-protected files
Key Features of PyMuPDF library:
- Fast processing (3-5x faster than alternatives) of large size PDF files
- High accuracy on complex document layouts
- Support for rich metadata extraction
Text extraction from a Web Page (web scraping using BeautifulSoup4 – a Python library for parsing HTML and XML documents):
- Parse HTML structure and remove boilerplate (scripts, styles, navigation, ads) from it
- Extract main content area (article, main tag, or heuristics)
- Parse meta tags (author, description, keywords) added to a web page
- Clean and normalize extracted text
Key Features of BeautifulSoup4 library:
- Intelligent content detection
- Metadata extraction from HTML meta tags
- Handles various HTML structures, including ‘messy’ HTML pages
There are still common Challenges:
- JavaScript-heavy sites may require headless browser
- Some sites have anti-scraping measures
- Content structure varies widely across sites
Stage 3: Extracted Text Chunking
Text extracted from a document must be split into manageable chunks to allow both LLM processing and embedding generation. Adopted chunking strategy significantly impacts system performance and accuracy.
In GraphRAG repository we use a Token-Based Chunking Strategy:
- Use BPE (Byte Pair Emcoding) tokenizer to convert text into tokens and then split it into chunks based on tokens count (not characters) for precise control over chunk size
- Default chunk size is 1024 tokens (~700-800 words)
- Chunks are created with a 15% chunk size overlap
- Preserve semantic boundaries are used for splitting wherever possible
Why Token-Based Chunking?:
- Data Type Compatibility: Embeddings API and LLM work with tokens, not characters
- Consistent Size: Character-based chunking can produce variable token counts per chunkl
- Semantic Preservation: Token boundaries often align with word boundaries
Overlap Rationale: 15% overlap ensures context isn’t lost at chunk boundaries. A sentence split between chunks will appear complete in at least one chunk.
Chunk ID Generation: Generate unique ID for a chunk using: MD5 hash (doc_id + text + chunk_index + offset). Including the document ID is critical as it prevents collisions when multiple documents contain similar content.
Stage 4: Entity Extraction (Multi-Pass Pipeline)
Entity extraction is the most sophisticated part of the document ingestion pipeline. The challenge is to accurately identify document authors and organizations they worked in at the time article was written separately from people or organizations mentioned in the document text. The pipeline logic is based on the position in a document of the text where authors and their organizations are typically mentioned”
- In a scientific article (PDF files that solution targets), both Author names and their Organizations are typically fount in the article header i.e., at the very beginning of the first chunk in a document
- In a blog or news publication (web pages that solution targets), Author names and possibly, their Organizations can be found either in the article header, or footer i.e., at the bottom of the last chunk in a document.
To address this challenge, the following multi-pass pipeline was implemented for the named entity extraction from a document added to the repository:
- PASS 1: Use Claude AI to extract entities of Person, Organization, Location, Technology, Concept, Product and Event types from all chunks in a document
- PASS 2: Use Claude AI to extract Author and their Organization entities:
- PDF file: Extract authors and their organization from the first 800 characters in the first chunk
- Web page: Extract authors and their organizations from the first 500 characters in the first chunk. If not found, try last 500 characters in the last chunk.
- Fallback to using document metadata, if PASS 2 extraction fails.
- PASS 3: Use Claude AI to extract semantic relationships between entities e.g., Author A collaborated with Person B or Organization C uses Technology T.
- PASS 4: Use statistical proximity analysis to define ‘mentioned with’ relationship between entities that appear frequently together in a document.
After all 4 passes are successfully completed, create corresponding nodes and edges (define relationship) in the Knowledge Graph (Neo4j). Rollback entity and relationship creation if any pass failed due to an error to prevent creating semantic inconsistencies in the graph.
PASS 1: General Entity Extraction
All text chunks in a document are processed by sending them in parallel (3-5 chunks at a time) to Claude AI with a custom prompt tailored for named entities. extraction. Custom prompt contains detailed instructions explaining Claude what entities should be extracted from the text. To help with identifying entities, it has several examples of text with entities targeted for extraction. Prompt also describes the format of expected response – JSON structure with entity name, type, confidence score (0.0 – 1.0), and description.
The following Entity Types are currently supported for extraction:
- Person: People mentioned in the chunk text
- Organization: Companies, institutions, or universities
- Location: Cities, countries, regions
- Technology: Frameworks, programming languages, algorithms
- Concept: Theoretical ideas, methodologies, approaches
- Product: Software products, platforms, services
- Event: Conferences, releases, historical events
De-duplication: after entities extraction from all chunks is completed, found entities are de-duplicated by normalizing their names (e.g., GPT-4″, “gpt-4”, “GPT 4” -> all become “gpt-4”). Only entities with highest confidence score are kept.
PASS 2: Specialized Author & Their Organization Extraction
Author extraction has a multi-step pipeline. Step 1: first chunk in a document is sent to Claude AI with a custom prompt tailored for extracting Authors (i.e., person who wrote the document) and Organizations they worked at when article was written. Apart from entity description and extraction instructions, prompt contains several text snippets showing how authors and their organization can appear in different document. Prompt uses the same response format as for other Entities – JSON structure with entity name, type, confidence score (0.0 – 1.0), and description.
To help Claude with locating authors and organizations they worked in, only first 800 characters (configurable) are sent for processing.
Step 2: if Step 1 didn’t find authors for a blog or an article published on a web page, pipeline tries extracting authors and their organizations from the last 500 characters of the last chunk in the document. The reason for doing that is that web publications often place authors right after the blog or article content.
Fallback to Document Metadata
If neither Step 1, nor Step 2 returned any authors, pipeline process makes “last resort” attempt to extract authors and their organization from PDF file metadata or web page meta-tags (OG, Twitter, etc).
The following Confidence Scores are assigned to extracted Author and Organization entities depending on the PASS:
- Step 1 or 2: Text extraction: 0.90-0.99 (high confidence)
- Step 3: Metadata extraction: 0.70-0.85 (medium confidence)
- Fallback: Using Unknown tag: 0.50 (low confidence)
Why Create “Unknown” Author?: Every document must have an author for relationship consistency.
PASS 3: Semantic Relationships
After extraction of supported Entities and Authors is completed, pipeline process proceeds to extracting semantic relationships from the document chunks. Relationships are extracted using Claude AI request with a highly-structured and constraints-based prompt.
The following Semantic Relationship types are extracted from a document:
RELATIONSHIP_TYPES = {
"WORKED_AT": {
"description": "Person/Author worked at Organization",
"source_types": ["Person", "Author"],
"target_types": ["Organization"],
"symmetric": False
},
"RELATED_TO": {
"description": "General semantic relationship between entities",
"source_types": ["*"], # Any entity type
"target_types": ["*"],
"symmetric": True,
"properties": ["context", "relationship_type", "confidence"]
},
"LOCATED_IN": {
"description": "Entity is physically located in a Location",
"source_types": ["Person", "Organization", "Event", "Product"],
"target_types": ["Location"],
"symmetric": False,
"properties": ["start_date", "end_date", "context"]
},
"COLLABORATED_WITH": {
"description": "Collaboration between authors or persons",
"source_types": ["Author", "Person"],
"target_types": ["Author", "Person"],
"symmetric": True,
"properties": ["project", "document_id", "confidence"]
},
"PART_OF": {
"description": "Hierarchical membership or composition",
"source_types": ["Person", "Organization", "Location", "Concept"],
"target_types": ["Organization", "Location", "Concept"],
"symmetric": False,
"properties": ["role", "context"]
},
"CREATED": {
"description": "Entity created another entity (product, technology)",
"source_types": ["Person", "Organization"],
"target_types": ["Product", "Technology", "Concept"],
"symmetric": False,
"properties": ["date", "context"]
},
"USES": {
"description": "Entity uses technology or product",
"source_types": ["Person", "Organization"],
"target_types": ["Technology", "Product"],
"symmetric": False
},
"IMPLEMENTS": {
"description": "Technology implements concept",
"source_types": ["Technology", "Product"],
"target_types": ["Concept"],
"symmetric": False
},
"PARTICIPATED_IN": {
"description": "Entity participated in an event",
"source_types": ["Person", "Organization", "Author"],
"target_types": ["Event"],
"symmetric": False,
"properties": ["role", "context"]
},
"OCCURRED_IN": {
"description": "Event occurred in location",
"source_types": ["Event"],
"target_types": ["Location"],
"symmetric": False,
"properties": ["date", "context"]
},
"AFFILIATED_WITH": {
"description": "Professional affiliation",
"source_types": ["Person", "Author"],
"target_types": ["Organization"],
"symmetric": False,
"properties": ["role", "start_date", "end_date"]
},
"MENTIONED_WITH": {
"description": "Co-occurrence in same context (chunk)",
"source_types": ["*"],
"target_types": ["*"],
"symmetric": True,
"properties": ["chunk_id", "frequency", "confidence"]
},
"FOUNDED": {
"description": "Person founded organization",
"source_types": ["Person", "Author"],
"target_types": ["Organization"],
"symmetric": False,
"properties": ["date", "context"]
}
}PASS 4: Co-occurrence Relationships
In PASS 4 entities are first grouped by chunk to identify which entities appear in each text chunk and to create pairwise relationships for every pair of entities that appear in the same chunk. A confidence score is calculated for each pair based on the individual entity confidence scores. For pairs with high confidence score MENTIONED_WITH relationship for a corresponding nodes is defined in Neo4j.
The co-occurrence relationship creates a layer in the knowledge graph that can be used for graph traversal, clustering, or as input for more advanced relationship inference.
Multi-Pass System Benefits
Accuracy: Separating author and their organization extraction from general entity extraction dramatically improves precision. Authors are identified based on document structure (headers or footers) rather than being confused with people emtioned in the document.
Confidence Tracking: Different extraction methods have different confidence levels, enabling confidence-based filtering and ranking.
Stage 5: Knowledge Graph Update
Step 1: Create Document and Chunk nodes
For each document added to repository a corresponding Document node is added to the Knowledge Graph (.Neo4j). Document title, source type (PDF or webpage), list of author names, etc. are stored as the node properties.
For each chunk generated for the document, a Chunk node with text content and metadata is added to the Graph. The following relationships are defined between Document node and Chunk nodes:
- (Document)–[:CONTAINS]–>(Chunk 1), (Chunk 2), …, (Last Chunk)
- (Chunk 1)–[:NEXT_CHUNK]–>(Chunk 2)–[NEXT_CHUNK]–> … –>(Last Chunk)
Step 2: Create Entity Nodes
Create or merge Entity nodes for each entity type (Person, Organization, Location, Technology, Concept, Product, and Event) extracted form the document.The following relationships are defined between Document, Chunk and Entity nodes:
- (Entity)–[:APPEARS_IN]–>(Document)
- (Entity)–[:MENTIONED_IN]–>(Chunk) for all chunks it is found.in.
Step 3: Create Author Nodes
Create or merge Author and Entity nodes of Organization type extracted from the document in the PASS 2 of extraction pipeline. The following relationships are defined between the nodes:
- (Document) — [:AUTHORED_BY] –> (Author)
- (Author) — [:WORKED_AT] –> (Organization)
- (Author)–[:APPEARS_IN]–>(Document)
- (Author)–[:MENTIONED_IN]–>(Chunk)
Step 4: Define Semantic Relationships
For Author and Entities of 7 supported types extracted from the document create or merge discovered semantic relationships following relationship rules described above:
- (Entity) -[:RELATED_TO]-> (Entity)
- (Entity) -[:COLLABORATED_WITH]-> (Entity)
- (Entity) -[:LOCATED_IN]-> (Entity)
- (Entity) -[:CREATED]-> (Entity)
- (Entity) -[:USES]-> (Entity)
- (Entity) -[:PART_OF]-> (Entity)
- (Entity) -[:AFFILIATED_WITH]-> (Entity)
- (Entity) -[:WORKED_AT]-> (Entity)
- (Entity) -[:COMPETED_WITH]-> (Entity)
- (Entity) -[:ACQUIRED]-> (Entity)
- (Entity) -[:FUNDED]-> (Entity)
Step 5: Create Entity Co-occurrence Relationships
For Author and Entities of 7 supported types extracted from the document create or merge co-occurrence relationships found in PASS 4 extraction with high confidence:
- (Entity) -[:MENTIONED_WITH]-> (Entity)
Note: Nodes and relationships are stored in Neo4j using MERGE (not CREATE) to ensure idem-potency and allow reprocessing documents without constraint violations.
Below is an example of Document, Chunks, Author and Entity nodes and edges/relationships created in Neo4j for an article Anthropic scientists hacked Claude’s brain — and it noticed. Here’s why that’s huge published by Venture Beat:
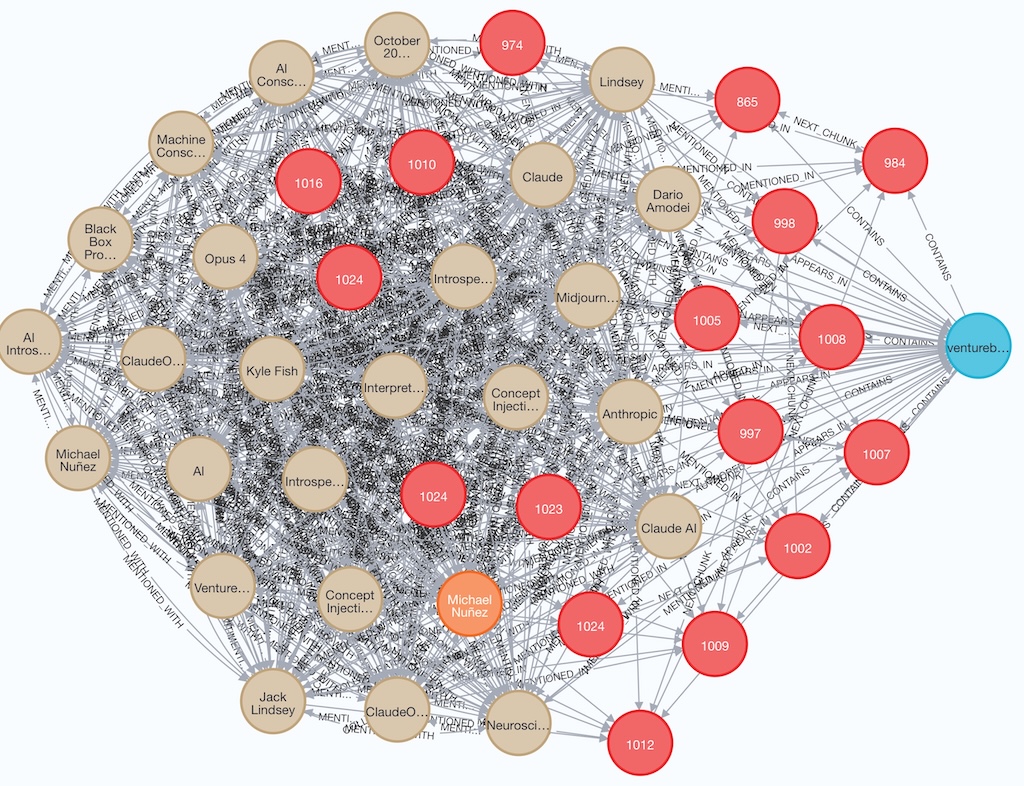
Stage 6: Generate Embeddings
At this pipeline stage embedding vectors are generated for the plain text in each chunk that was stored in Neo4j for submitted document and then stored them in vector database Chroma.
- Step 1: retrieve chunks with meaningful content (i.e., chunks with non-zero tokens count) from Neo4j for processing
- Step2: check if embeddings for a chunk already exists in Chroma DB using plain text SHA-256 hash and skip unchanged chunks.
- Step 3: processes chunks in batches for efficiency usingVoyage AI (“voyage-3” model) to generate embeddings vector with 1024 dimensions.
- Step 4: store embedding in Chrome DB collection in the following format
{
"ids": ["doc_id:chunk_index"], # Unique identifiers
"embeddings": [[1024 float values]], # voyage-3 vectors
"metadatas": [{
"doc_id": "uuid",
"chunk_index": int,
"hash": "sha256_hash", # For change detection
"token_count": int,
"created_at": "timestamp"
}],
"documents": ["chunk text content"] # Full text for retrieval
}GraphRAG Server REST API Design
GraphRAG document repository server application exposes three categories of APIs – health monitoring, documents ingestion & management, and search.
Health & System Monitoring API
- GET /api/health – Basic health check
- GET /api/status – Comprehensive system status with database statistics
Documents Ingestion & Management API
Core APIs
- POST /api/documents/upload – upload a PDF file to the document repository
Sample API response:
{
"success": true,
"document": {
"id": "5994e9a3-169d-4db0-8f2a-6a2bba124498",
"filename": "2210.03629v3.pdf",
"object_name": "5994e9a3-169d-4db0-8f2a-6a2bba124498/2210.03629v3.pdf",
"hash": "f285b0971ae4a790e402fb93966bed3adde2cf0a04977d08b2b40d6ab0cace69",
"size": 633805,
"content_type": "application/pdf",
"source_type": "pdf",
"presigned_url": "http://localhost:9000/documents/5994e9a3-169d-4db0-8f2a-6a2bba124498/2210.03629v3.pdf?X-Amz-Algorithm=AWS4-HMAC-SHA256&X-Amz-Credential=minioadmin%2F20251116%2Fus-east-1%2Fs3%2Faws4_request&X-Amz-Date=20251116T010358Z&X-Amz-Expires=86400&X-Amz-SignedHeaders=host&X-Amz-Signature=49d1ebf8c112da411b8e0c5511b894336f9b7df10af6be65fc56d98ccf044390",
"metadata": {
"doc_id": "5994e9a3-169d-4db0-8f2a-6a2bba124498",
"original_filename": "2210.03629v3.pdf",
"source_type": "pdf",
"file_hash": "f285b0971ae4a790e402fb93966bed3adde2cf0a04977d08b2b40d6ab0cace69",
"upload_timestamp": "2025-11-16T01:03:58.598796",
"processing": {
"status": "completed",
"chunks_created": 39,
"processing_time": 1.276457,
"text_statistics": {
"page_count": 33,
"total_characters": 110319,
"total_words": 17108,
"pages_with_text": 33
},
"graph_storage": {
"status": "completed",
"document_stored": true,
"chunks_stored": true,
"chunks_count": 39
},
"embeddings": {
"status": "completed",
"chunks_embedded": 39,
"embeddings_stored": 39,
"processing_time": 2.3364198207855225,
"model": "voyage-3",
"dimensions": 1024,
"metadata": {
"model": "voyage-3",
"usage": {
"total_tokens": 1950
},
"embedding_dimensions": 1024,
"chroma_collection": "document_chunks"
}
}
}
},
"created_at": "2025-11-16T01:03:58.608947"
},
"message": "Document uploaded and processed successfully",
"duplicate": false
}- POST /api/documents/add-url – add a document by fetching content from a URL
Sample API response:
{
"success": true,
"document": {
"id": "7773f9a8-5eaf-41ca-9048-608a36903bea",
"filename": "venturebeat.com_9471f509-13ef-4dcc-aa88-e919618e640c.txt",
"object_name": "7773f9a8-5eaf-41ca-9048-608a36903bea/venturebeat.com_9471f509-13ef-4dcc-aa88-e919618e640c.txt",
"hash": "60c0523381abb79057e94e73b9d2d51654e290503fed2f2ca73625f4f64d42c5",
"size": 43211,
"content_type": "application/octet-stream",
"source_type": "webpage",
"presigned_url": "http://localhost:9000/documents/7773f9a8-5eaf-41ca-9048-608a36903bea/venturebeat.com_9471f509-13ef-4dcc-aa88-e919618e640c.txt?X-Amz-Algorithm=AWS4-HMAC-SHA256&X-Amz-Credential=minioadmin%2F20251116%2Fus-east-1%2Fs3%2Faws4_request&X-Amz-Date=20251116T003858Z&X-Amz-Expires=86400&X-Amz-SignedHeaders=host&X-Amz-Signature=a59fb6f82e104409fc149dcffb7824ea0417436fefd2ed71000b0f260c69fc74",
"metadata": {
"doc_id": "7773f9a8-5eaf-41ca-9048-608a36903bea",
"original_filename": "venturebeat.com_9471f509-13ef-4dcc-aa88-e919618e640c.txt",
"source_type": "webpage",
"file_hash": "60c0523381abb79057e94e73b9d2d51654e290503fed2f2ca73625f4f64d42c5",
"upload_timestamp": "2025-11-16T00:38:58.906093",
"title": "Databricks: 'PDF parsing for agentic AI is still unsolved' — new tool replaces multi-service pipelines with single function | VentureBeat",
"source_url": "https://venturebeat.com/data-infrastructure/databricks-pdf-parsing-for-agentic-ai-is-still-unsolved-new-tool-replaces",
"og_title": "Databricks: 'PDF parsing for agentic AI is still unsolved' — new tool replaces multi-service pipelines with single function",
"twitter_title": "Databricks: 'PDF parsing for agentic AI is still unsolved' — new tool replaces multi-service pipelines with single function",
"twitter_creator": "@venturebeat",
"article_published_time": "2025-11-14T11:00-05:00",
"language": "en",
"content_type": "text/html; charset=utf-8",
"last_modified": "",
"server": "Vercel",
"domain": "venturebeat.com",
"scheme": "https",
"final_url": "venturebeat.com",
"scraped_at": "2025-11-16T00:36:18.233072",
"processing": {
"status": "completed",
"chunks_created": 9,
"processing_time": 0.893602,
"text_statistics": {
"total_characters": 43169,
"total_words": 5713,
"response_size": 132416,
"status_code": 200
},
"graph_storage": {
"status": "completed",
"document_stored": true,
"chunks_stored": true,
"chunks_count": 9
},
"embeddings": {
"status": "completed",
"chunks_embedded": 9,
"embeddings_stored": 9,
"processing_time": 0.9918451309204102,
"model": "voyage-3",
"dimensions": 1024,
"metadata": {
"model": "voyage-3",
"usage": {
"total_tokens": 450
},
"embedding_dimensions": 1024,
"chroma_collection": "document_chunks"
}
},
"url": "https://venturebeat.com/data-infrastructure/databricks-pdf-parsing-for-agentic-ai-is-still-unsolved-new-tool-replaces"
}
},
"created_at": "2025-11-16T00:38:58.910172"
},
"message": "Webpage processed and stored successfully",
"duplicate": false
}
- GET /api/documents – list document in repository with pagination / filtering support
- GET /api/documents/{doc_id} – get metadata for a given document
- DELETE /api/documents/{doc_id} – delete a document and all associated data from Neo4j, Chroma DB, and MinIO object storage
- GET /api/documents/{doc_id}/authors – retrieve all authors for a given document
- GET /api/documents/{doc_id}/chunks – get all text chunks for a a given document from graph DB (Neo4j)
- POST /api/documents/search/similar – find chunks matching a query using vector similarity
- GET /api/documents/authors – search for authors with optional filtering and pagination
- GET /api/documents/authors/{author_name}/documents – retrieve all documents authored by a given author
- POST /api/documents/{doc_id}/embeddings – generate or regenerate embeddings for all chunks of a document
- GET /api/documents{doc_id}/chunk-embeddings – get detailed information about chunks with embeddings in Chroma DB
- POST /api/documents/{doc_id/entities – extract entities from a document using Claude API
- GET /api/documents/{doc_id}/entities – retrieve all entities extracted from a document
- GET /api/documents/entities/types/{entity_type} – find entities of a specific type with pagination support
- GET /api/documents/entities/{entity_id} – get detailed information about a specific entity
- GET /api/documents/entities/{entity_id}/related – find entities related to a given entity through co-occurrence
- GET /api/documents/relationships/types – Get all defined relationship types and their constraints
- POST /api/documents/{doc_id}/relationships/extract – extract semantic relationships between entities in a specific document
- GET /api/documents/entities/{entity_id/relationships – get all relationships for a specific entity with optional filtering
- GET /api/documents/{doc_id}/relationships – get all relationships extracted from a specific document
Batch APIs
- POST /api/documents/batch/embeddings – generate embeddings for a batch of documents
- POST /api/documents/batch/entities – extract entities from multiple documents in batch
Debug and Statistics APIs
- GET /api/documents/graph/statistics – get statistics about the knowledge graph data (documents, chunks, relationships)
- GET /api/documents/authors/statistics – get statistics about authors in the system
- GET /api/documents/embeddings/statistics – get statistics about embeddings across all documents in repository
- GET /api/documents/entities/statistics – get statistics about extracted entities across all documents
- GET /api/documents/relationships/statistics – get statistics about all relationships in the knowledge graph
- GET /api/documents/{doc_id}/diagnostics – debug endpoint to check document-chunk-entity relationships in Neo4j
Search API
Core Search APIs
- POST /api/search/query – main search endpoint supporting multiple search modes.
- Vector search: semantic similarity using embeddings
- Graph search: entity-based traversal and relationships
- Hybrid search: combination of vector and graph approaches
- GET /api/search/modes – get available search modes and their descriptions.
- POST /api/search/explain – analyze a query and provide explanation with mode suggestion
- Query complexity assessment
- Detected entities and concepts
- Recommended search mode
- Suggested mode weights for hybrid search
Search Statistics and Analytics APIs
- GET /api/search/statistics – get search system statistics and analytics
- GET /api/search/analytics/summary – get detailed analytics summary with trends and insights.
- GET /api/search/analytics/trends – get trending queries and search patterns.
- GET /api/search/analytics/performance – get performance insights and optimization recommendations.
- GET /api/search/health – health check for search functionality.
Conclusion
In Part 1 of the series of blogs about GraphRAG document repository system I covered:
- Multi Database Architecture: Neo4j knowledge graph for document and entity relationships , Chroma DB for semantic search, MinIO for file storage – each optimized for its specific purpose.
- Intelligent Document Processing Pipeline: automatic document ingestion pipeline to transform raw PDF files or text extracted from a web page into rich knowledge graphs with semantic embeddings.
- Multi Pass Entity Extraction: AI-powered named entities and their relationships extraction from documents added to the repository.
- Knowledge Graph Model: Document centric schema with 7 entity types, optimized relationship directions, and sequential chunk linking.
- Documents and Search APIs: core back-end server APIs for Health document ingestion and multi-mode (graph, vector and hybrid) search.
In subsequent parts of the blog I will talk about advanced AI-powered entity and relationship extraction from repository corpus, including global entity co-occurrence, cross-document relation extraction, and communities generation and then about, AI-powered document query generation in response to a natural-language user question. Stay tuned.


2 Comments